 | |||
| Home | Search | Reports | Help |
This page offers a demonstration of the use of DracoBase. It is intended for developers.
Please contact Paul Szauter with comments and questions.
Outline of DNA to Trait Projects
This is adapted from the shared document Outline of DNA to Trait Projects, April 2011.
| DNA to Trait 3a: bog breath |
DNA to Trait 3. Identifying the gene behind bog breath ( bog or Otc).
Students will first encounter bog breath as a sex-linked trait that is basically a conditional lethal. On the Celestial Diet (rice, vegetables, sesame seeds and herbs), homozygous females and hemizygous males are fine. If they are fed the standard diet (meat and bones) they get very sick and may die. Drakes that are sick with bog breath have a noticeable odor of ammonia in their breath (hence the name).
There are some low-level investigations of the phenotype that would be informative. These are not crosses, and are not supported by the current game screens. At the simplest level, information could be fed to students as short reports using text and images.
First, an investigation of the Celestial Diet will reveal that it is basically a low-protein diet supplemented with arginine (sesame seeds are high in arginine). The very expensive mixture of herbs is in the diet for "magical" reasons and contains nothing of importance. It is possible to produce a cheap rice + vegetables + arginine diet that will permit these drakes to survive. Omission of the arginine will cause stunted growth. Again, it is not necessary for students to confirm this; they can simply read the results of a study.
Second, an investigation of the blood chemistry of drakes undergoing an episode of bog breath will reveal hyperammonemia (a high level of ammonia in the blood). This is broadly indicative of a disorder in the metabolism of waste nitrogen derived from the catabolism of amino acids. Again, students could get this information as a report.
It would be a bit much to expect students to connect the dots here, so a fictitious mentor in the game might suggest investigating inherited disorders of the urea cycle. There are five enzymes in this metabolic cycle as shown below. There is also an important transporter gene. The urea cycle takes in waste nitrogen as ammonia and synthesizes arginine as a byproduct. Drakes with bog breath need an arginine supplement because, for them, it is an essential amino acid, one that they cannot synthesize.
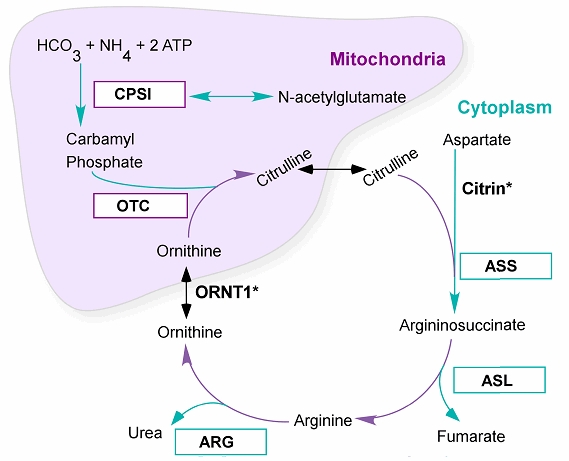
| Symbol | Name | Function |
| Cps1 | Carbamoyl-phosphate synthetase 1 | 2 ATP + NH3 + CO2 + H2O = 2 ADP + phosphate + carbamoyl phosphate |
| Slc25a15 (ORNT1) | Ornithine transporter | Ornithine transport across inner mitochondrial membrane, from the cytoplasm to the matrix |
| Otc | Ornithine carbamoyltransferase | Carbamoyl phosphate + L-ornithine = phosphate + L-citrulline |
| Ass1 | Argininosuccinate synthetase 1 | ATP + L-citrulline + L-aspartate = AMP + diphosphate + N(omega)-(L-arginino)succinate |
| Asl | Argininosuccinate lyase | 2-(N(omega)-L-arginino)succinate = fumarate + L-arginine |
| Arg | Arginase | L-arginine + H2O = L-ornithine + urea |
This is enough background to consider approaches to identifying the gene involved. The exact approach used in the game depends on our decision on the path to the introduction of DNA sequences for drakes. It is possible to imagine a low-cost strategy of isolating a gene at a time, but if Geniverse is following contemporary genomics, it might be a better approach to go from no sequences at all to a complete genome assembly with some functional annotation of most genes.
Imagine that shortly after the information on bog breath as a urea cycle disorder is conveyed to students, the drake genome sequence becomes available. There is some kind of a low-tech database available to students.
At this point, there are two approaches. The elegant approach is to ask whether any of the five genes for urea cycle enzymes is sex linked. Only the ornithine transcarbamylase ( Otc) gene is on the X chromosome. The other approach is to investigate all five urea cycle enzymes and the transporter.
The investigation consists of sequencing the Otc gene (or all six genes) from drakes known to carry bog breath. Students would use a simple database tool to compare the predicted amino acid sequence from the mutants with the standard sequence from a wild-type strain (e.g. Nightwing).
For the actual mutation, we would simply steal the nonconservative amino acid substitution from the human cases documented in OMIM. Here is a good citation from the OMIM entry. We would use mutations associated with "late onset" type mutations, which tend to alter amino acids on the protein surface rather than in the active site:
http://pubget.com/paper/8544185
From this outline, you can see that this requires us to: 1) devise ways to feed research information to students (easy), 2) build a genomic database that supports simple queries, and 3) devise (or steal) a tool that allows the comparison of DNA and amino acid sequences.
Let us imagine that we have got students to the point that they are ready to take a drake with bog breath from their stable and send a DNA sample to a facility for sequencing. They ask that the Otc gene be sequenced. If they have more gold than wit, they can have all six genes sequenced. The computational biology and genomics in Geniverse are better than what we have in our world. The facilty returns the results to the student in the form of links.
The links go to pages that display the sequence of the predicted transcript(s) from the mutant drake. This is hard to interpret by itself, but such is the power of steampunk genomics that the sequence from the mutant drake is aligned to the standard sequence from Nightwing. In addition, a predicted protein sequence is generated and aligned to the standard sequence from Nightwing.
What would such pages look like? Click the links below.
Otc sequence from bog breath Drake
Cps1 sequence from bog breath Drake
Slc25a15 sequence from bog breath Drake
Ass1 sequence from bog breath Drake
Asl sequence from bog breath Drake
Arg sequence from bog breath Drake
| DNA to Trait 3b: dancer |
DNA to Trait 3. Identifying the gene behind dancer ( dcr or Slc25a15). As long as we are looking at urea cycle disorders, let's consider dancer. Investigation of the phenotype can be similar to that used for bog breath as outlined above. When it comes time to sequence genes, the best students might take this as a mapping challenge, mapping the dancer phenotype to a region with a limited number of candidate genes. The Slc25a15 gene is going to stand out as highly relevant. If they have more gold than wit, they can just sequence all six genes as above. Here are the results from sequencing all six genes from a drake afflicted with dancer. Slc25a15 sequence from dancer Drake Cps1 sequence from dancer Drake Otc sequence from dancer Drake Ass1 sequence from dancer Drake |